9.5: Recommended standards for the reduction of bacterial contamination of blood components
In recent years bacterial contamination of blood has been significantly reduced by the introduction of improved donor arm cleansing using 70% isopropyl alcohol/2% chlorhexidine gluconate applied as a single-step procedure, and diversion of the first 20–30 mL of the blood donation. The risk of bacterial contamination can be further reduced, but not eliminated, by screening of blood components.
9.5.1: Arm cleansing
There should be an effective, specified and validated method of arm cleansing, using an approved skin-cleansing system. 70% isopropyl alcohol/2% chlorhexidine gluconate is recommended by the National Evidence-Based Guidelines for Preventing Healthcare-Associated Infections in NHS Hospitals in England4. Adherence to the principles, protocols and practices relating to the correct use of the specified skin-cleansing system shall be regularly audited by periodic observation and corrected if found to be lacking.
9.5.2: Diversion of donation
A minimum of 20 mL of the first part of every blood donation should be diverted into a side-arm pouch, in order to minimise the level of bacterial skin contaminants in the collection bag. This diverted volume can be used as a source of blood samples for mandatory and other testing of the donation.
9.5.3: Screening of platelet components
There should be a means of detecting bacterial contamination of platelet components, using validated methods. The key requirements of a detection system are (i) effective sample size, (ii) a rapid test result or automated continuous monitoring with alarm notification and (iii) reliable detection of bacteria at a level indicating potential risk to recipient.
Bacterial culture using an automated microbial detection system represents the most widely used and efficient method for screening of components. Its key feature is the continuous monitoring of incubated culture bottles to allow immediate withdrawal of contaminated and associated units.
The use of an automated microbial detection system using the following protocol has been shown to give a substantial risk reduction regarding transfusion transmission of bacteria in platelet components5. This requires a minimum hold period of at least 36 hours before sampling and a minimum of 8 mL inoculated into both anaerobic and aerobic culture bottles. Continuous incubation and monitoring need to be performed until the end of shelf life, which can be extended from 5 to 7 days.
9.5.3.1: Single-test protocol
1. Platelet components are held for at least 36 hours after collection
2. Minimum 8 mL samples are inoculated into each aerobic and anaerobic bottle.
3. If samples are negative after a minimum of 6 hours of incubation, release product on a negative-to-date basis with 7-day shelf life and continue incubation and monitoring for the shelf life of the product.
4. A protocol must be in place for confirmation of the presence of contamination.
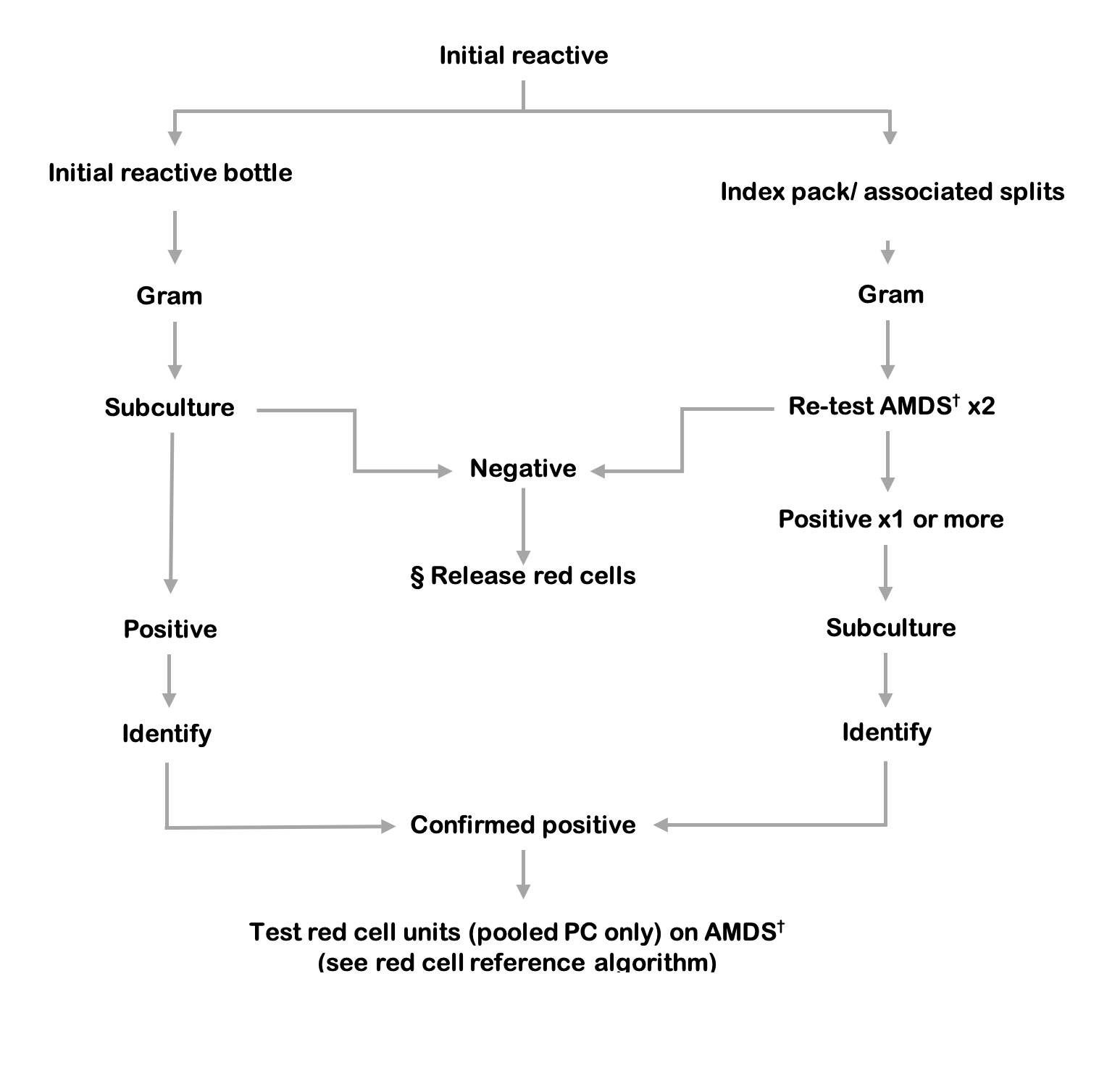
Figure 9.6 Platelet components testing algorithm. If the index pooled platelet component is not available to re-test, the associated red cell unit must be tested.
§ Release of red cells requires a negative result from both the index culture bottles and testing of the platelet component.
† AMDS: Automated microbial detection system, re-test aerobic and anaerobic culture in duplicate.
The following definitions of screening test results are recommended.
Initial reactive: Index culture bottle(s), from initial screening, with positive signal from an automated microbial detection system.
Repeat reactive: Repeat culture bottle(s), from repeat sampling of the index unit, with positive signal from an automated microbial detection system.
Associated reactive: Associated culture bottle(s), from sampling of associated components, with positive signal from an automated microbial detection system.
Confirmed positive: Matching speciation from positive subculture of the initial reactive AND the repeat reactive OR the associated reactive.
Indeterminate positive: A combination of results that includes positive subculture but does not satisfy the definition for ‘confirmed positive’.
- Positive subculture from the initial reactive, BUT no positive signal from sampling of the index or associated components OR no index or associated components returned for sampling.
- Positive subculture from ONLY ONE of the initial reactive OR the repeat reactive OR the associated reactive.
- Non-matching speciation from positive subcultures of the initial reactive AND the repeat reactive OR the associated reactive.
(In most cases there is positive subculture from the initial reactive bottle but the index unit is not available for culture because it has been transfused).
Indeterminate negative: Negative subculture from the initial reactive, BUT no index unit available for testing.
(Where assessed, negativity would also require no organisms on Gram stain and negative growth curves from the automated microbial detection system).
False positive: Negative subculture from the initial reactive AND no signal from repeat sampling of the index unit.
False negative: Negative screening test to the end of the incubation period, BUT positive subculture from sampling of the platelet unit during investigation of a visually abnormal unit OR a post-transfusion reaction.
When the initial reactive result is generated, any associated components (plasma, red cells etc. from the same donation) must be quarantined pending the result of repeat sampling and a recall procedure should be initiated for any platelet units or other components already issued.
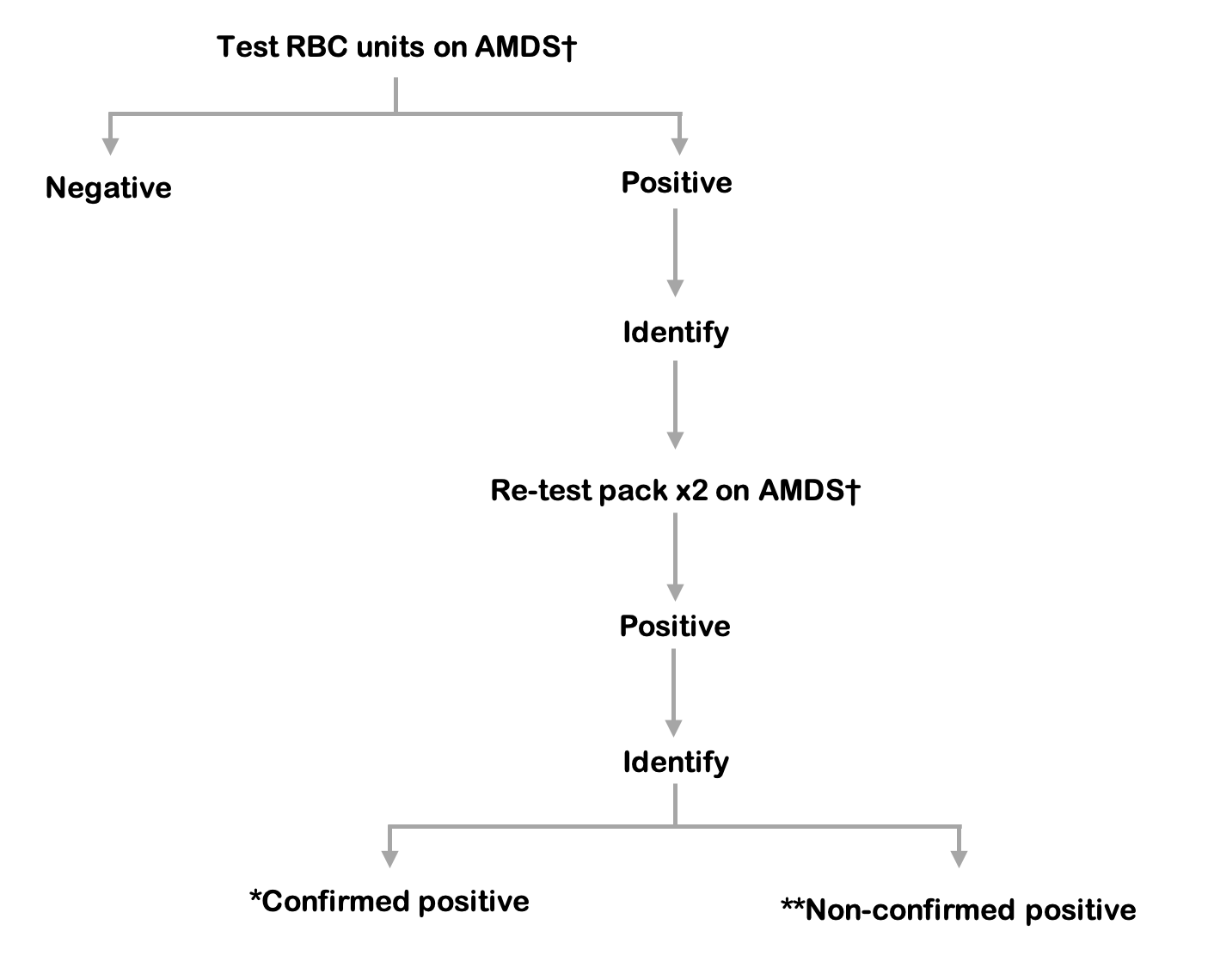
Figure 9.7 Red blood cell reference algorithm
* Confirmed positive: a match at species level on the initial RBC test and re-test.
** Non-confirmed positive: negative repeat test and no match at species level with the PC confirmed positive result.
† AMDS: Automated microbial detection system, re-test aerobic and anaerobic culture in duplicate.
Release of red cells requires a negative result from both the index culture bottles and testing of the platelet component.
Testing algorithms shown in Figures 9.6 and 9.7 are guidelines and may be modified to be Blood Service specific.